Laboratory Diagnosis
Since many mycoplasmas are fastidious, special attention must be taken on techniques for obtaining, transporting, and processing clinical specimens. Bacterial and fungal contamination as well as environmental conditions resulting in loss of viable mycoplasma organisms should be avoided. Samples from live animals are limited to body secretions and accessible sites amenable to swabbing. Swabs must be placed in an appropriate transport medium such as Amies' without charcoal or 2-SP. Joint fluid aspirates, milk samples, tracheal and bronchoalveolar washings as well as biopsies are further specimens appropriate for mycoplasma isolation. At necropsy samples of affected tissue with visible lesions should be taken for culture of mycoplasmas. In general, samples to be submitted for mycoplasma isolation must be kept moist and cool, and transported fast to the laboratory.
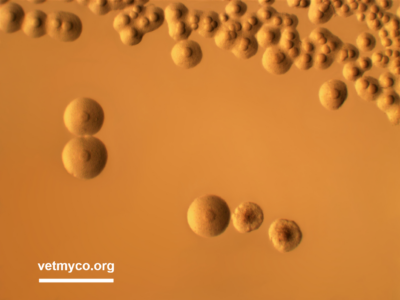
Laboratory diagnosis of mycoplasma infections is commonly achieved by conventional cultivation procedures by which mycoplasmas produce colonies with diameters of 0.1 mm (ureaplasmas) or between 0.2 and 1 mm (mycoplasmas, acholeplasmas) on agar medium, mostly with a characteristic fried egg morphology. Some Mycoplasma and Acholeplasma species, however, more likely produce centerless, granulated, or mulberry-like colonies, and differences in the colonies’ opacity may occur (Fig. 1). Once received at the laboratory, one part of the sample is usually plated directly on agar medium which allows quantification of mycoplasmas present in the samples while part is incubated in broth medium to increase the probability of isolation. Broth-to-broth passages increase isolation results since samples such as tissue homogenates may release mycoplasmacidal substances. Several complex media formulations are used for the isolation of mycoplasmas, most or all containing components such as beef heart infusion, peptone, yeast extract, animal sera, and various supplements. Well known media for the isolation of mycoplasmas include Eaton, Hayflick, Friis, Frey, and SP-4 (Mycoplasma, Acholeplasma) as well as ‘Ureaplasma’ and U4 (Ureaplasma) medium. Agar plates are usually incubated at 36-37°C under microaerobic conditions (5-7% CO2 atmosphere) for up to 14 days and daily checked for colony formation using a stereomicroscope. Broth cultures are incubated at 36-37°C in ambient air for 7-10 days or until a color change of the medium is observed, followed by subcultivation on agar plates. Cultivation procedures also include examination of a limited number of biochemical properties such as glucose fermentation and arginine or urea hydrolysis only enabling the assignment of an isolate to the genus Ureaplasma or a metabolic group within genus Mycoplasma.
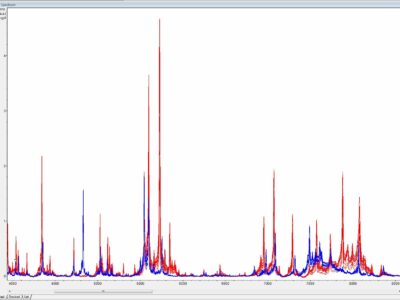
Once cultivated species identification of an isolate is usually achieved by antigenic or molecular methods. Antigenic identification tests depend on specific antisera to each individual mycoplasma species which are not readily available in most diagnostic laboratories. In addition, specific antisera may vary in their capacity to identify mycoplasmas because of multiple cross-reactions and substantial serological heterogeneity of some mycoplasma species. Molecular identification of mycoplasma isolates is based on species specific PCRs or more commonly on universal PCRs targeting the 16S rRNA gene, the 16S-23S intergenic spacer region (ISR) or the rpoB gene followed by sequencing or amplicon analyses (e.g. restriction fragment length polymorphism - RFLP). Nevertheless, both antigenic and molecular identification techniques are time-consuming, labor-intensive, not always discriminating, and require expertise which is rather restricted to specialized laboratories. Recently, MALDI-ToF mass spectrometry has been reported to be a superior tool for the identification and differentiation of animal mycoplasmas by combining convenience, speed, and precision, and thus complying with demands of modern diagnostic veterinary microbiology (Fig. 2).
Fig. 2. Substantial differences between overlaid MALDI-ToF mass spectra generated from closely related Mycoplasma bovis PG45T (in blue, n = 15) and Mycoplasma agalactiae PG2T (in red, n = 15), y-axis - a.u. = arbitrary unit, x-axis - m/z = mass-to-charge ratio. Credits: Joachim Spergser (Vetmeduni Vienna)
Numerous conventional and real-time PCR systems have been developed for species-specific detection of animal mycoplasmas in clinical specimens. Species-specific PCRs are commonly applied for the detection of uncultivable mycoplasmas (e.g. hemoplasmas), highly fastidious mycoplasma species (e.g. Mycoplasma hyopneumoniae) and mycoplasmas causing diseases of high veterinary importance. In addition, simultaneous detection of multiple mycoplasma species can be achieved by employing multiplex PCRs, PCR with denaturing gradient gel electrophoresis (DGGE) or DNA microarrays.
Only few serological assays for the detection of the host immune response to animal mycoplasmas have been standardized and made commercially available, so far. Immunoassay results are sometimes difficult to interpret poorly correlating with infection and disease, and thus several developed tests remain primarily research tools not applicable for routine diagnostic purposes at present. Nevertheless, immunoassays have shown to be useful as a herd test, and for demonstrating lack of infection.
The effectiveness of antimicrobials in vivo can be indirectly assessed by in vitro susceptibility testing to determine the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of an antimicrobial agent toward Mycoplasma strains. However, susceptibility tests for animal mycoplasmas have not been standardized so far and relating MIC results to effectiveness of the antimicrobials in use (commonly referred as clinical breakpoints) has not been determined. In consequence, results obtained from non-standardized susceptibility tests should be interpreted with caution and may only serve as guidance for choosing an antibiotic to treat a mycoplasma infection.
References
- Calcutt, M.J., Lysnyansky, I., Sachse, K., Fox, L.K., Nicholas, R.A.J. and R.D. Ayling. 2018. Gap analysis of Mycoplasma bovis disease, diagnosis and control: An aid to identify future development requirements. Transbound. Emerg. Dis. 65 Suppl. 1: 91-109.
- Gautier-Bouchardon, A.V. 2018. Antimicrobial resistance in Mycoplasma spp. Microbiol. Spectr. 6: 1–21.
- Markham, P.F., Noormohammadi, A.H. 2005. Diagnosis of mycoplasmosis in animals. In: Blanchard, A. and G. Browning (eds). Mycoplasmas: Molecular biology, pathogenicity and strategies for control. Horizon Bioscience, Norfolk, UK.
- McAuliffe, L., Ellis, R.J., Lawes, J.R., Ayling, R.D., Nicholas, R.A. 2005. 16S rDNA PCR and denaturing gradient gel electrophoresis; a single generic test for detecting and differentiating Mycoplasma species. J. Med. Microbiol. 54: 731–739.
- Razin, S., Yogev, D. and Y. Naot.1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62: 1094–1156.
- Spergser, J., Rosengarten, R. 2007. Identification and differentiation of canine Mycoplasma isolates by 16S-23S rDNA PCR-RFLP. Vet. Microbiol. 125: 170–174.
- Spergser, J., Hess, C., Loncaric, I. and A.S. Ramírez. 2019. Matrix-assisted laser desorption ionization-time of flight mass spectrometry is a superior diagnostic tool for the identification and differentiation of mycoplasmas isolated from animals. J. Clin. Microbiol. 57 (9), pii: e00316-19.
- Ramírez, A.S., Naylor, C.J., Pitcher, D.G., Bradbury, J.M. 2008. High inter-species and low intra-species variation in 16S-23S rDNA spacer sequences of pathogenic avian mycoplamas offers potential use as a diagnostic tool. Vet. Microbiol. 128: 279–287.
- Volokhov, D.V., Simonyan, V., Davidson, M.K., Chizhikov, V.E. 2012. RNA polymerase beta subunit (rpoB) gene and the 16S-23S rRNA intergenic transcribed spacer region (ITS) as complementary molecular marker in addition to the 16S rRNA gene for phylogenetic analysis and identification of the species of the family Mycoplasmataceae. Mol. Phylogenet. Evol. 62: 515–528
- Whitford, H.W. 1994. Isolation of mycoplasmas from clinical specimens. In: Whitford, H.W., Rosenbusch, R.F. and L.H. Lauerman (eds.). Mycoplasmosis in animals: Laboratory diagnosis. Iowa State University Press, USA